Advection-diffusion of tracer by cellular flow
This example can be viewed as a Jupyter notebook via 
An example demonstrating the advection-diffusion of a tracer by a cellular flow.
Install dependencies
First let's make sure we have all required packages installed.
using Pkg
pkg"add PassiveTracerFlows, CairoMakie, Printf"Let's begin
Let's load PassiveTracerFlows.jl and some other needed packages.
using PassiveTracerFlows, CairoMakie, PrintfChoosing a device: CPU or GPU
dev = CPU() # Device (CPU/GPU)Numerical parameters and time-stepping parameters
nx = 128 # 2D resolution = nx²
stepper = "RK4" # timestepper
dt = 0.02 # timestep
nsteps = 800 # total number of time-steps
nsubs = 25 # number of time-steps for intermediate logging/plotting (nsteps must be multiple of nsubs)Numerical parameters and time-stepping parameters
Lx = 2π # domain size
κ = 0.002 # diffusivitySet up cellular flow
We create a two-dimensional grid to construct the cellular flow. Our cellular flow is derived from a streamfunction $ψ(x, y) = ψ₀ \cos(x) \cos(y)$ as $(u, v) = (-∂_y ψ, ∂_x ψ)$. The cellular flow is then passed into the TwoDAdvectingFlow constructor with steadyflow = true to indicate that the flow is not time dependent.
grid = TwoDGrid(dev; nx, Lx)
ψ₀ = 0.2
mx, my = 1, 1
ψ = [ψ₀ * cos(mx * grid.x[i]) * cos(my * grid.y[j]) for i in 1:grid.nx, j in 1:grid.ny]
uvel(x, y) = ψ₀ * my * cos(mx * x) * sin(my * y)
vvel(x, y) = -ψ₀ * mx * sin(mx * x) * cos(my * y)
advecting_flow = TwoDAdvectingFlow(; u = uvel, v = vvel, steadyflow = true)Problem setup
We initialize a Problem by providing a set of keyword arguments.
prob = TracerAdvectionDiffusion.Problem(dev, advecting_flow; nx, Lx, κ, dt, stepper)and define some shortcuts
sol, clock, vars, params, grid = prob.sol, prob.clock, prob.vars, prob.params, prob.grid
x, y = grid.x, grid.ySetting initial conditions
Our initial condition for the tracer $c$ is a gaussian centered at $(x, y) = (L_x/5, 0)$.
gaussian(x, y, σ) = exp(-(x^2 + y^2) / (2σ^2))
amplitude, spread = 0.5, 0.15
c₀ = [amplitude * gaussian(x[i] - 0.2 * grid.Lx, y[j], spread) for i=1:grid.nx, j=1:grid.ny]
TracerAdvectionDiffusion.set_c!(prob, c₀)Time-stepping the Problem forward
We want to step the Problem forward in time and, whilst doing so, we'd like to produce an animation of the tracer concentration.
First we create a figure using Observables.
c_anim = Observable(Array(vars.c))
title = Observable(@sprintf("concentration, t = %.2f", clock.t))
Lx, Ly = grid.Lx, grid.Ly
fig = Figure(size = (600, 600))
ax = Axis(fig[1, 1],
xlabel = "x",
ylabel = "y",
aspect = 1,
title = title,
limits = ((-Lx/2, Lx/2), (-Ly/2, Ly/2)))
hm = heatmap!(ax, x, y, c_anim;
colormap = :balance, colorrange = (-0.2, 0.2))
contour!(ax, x, y, ψ;
levels = 0.0125:0.025:0.2, color = :grey, linestyle = :solid)
contour!(ax, x, y, ψ;
levels = -0.1875:0.025:-0.0125, color = (:grey, 0.8), linestyle = :dash)
fig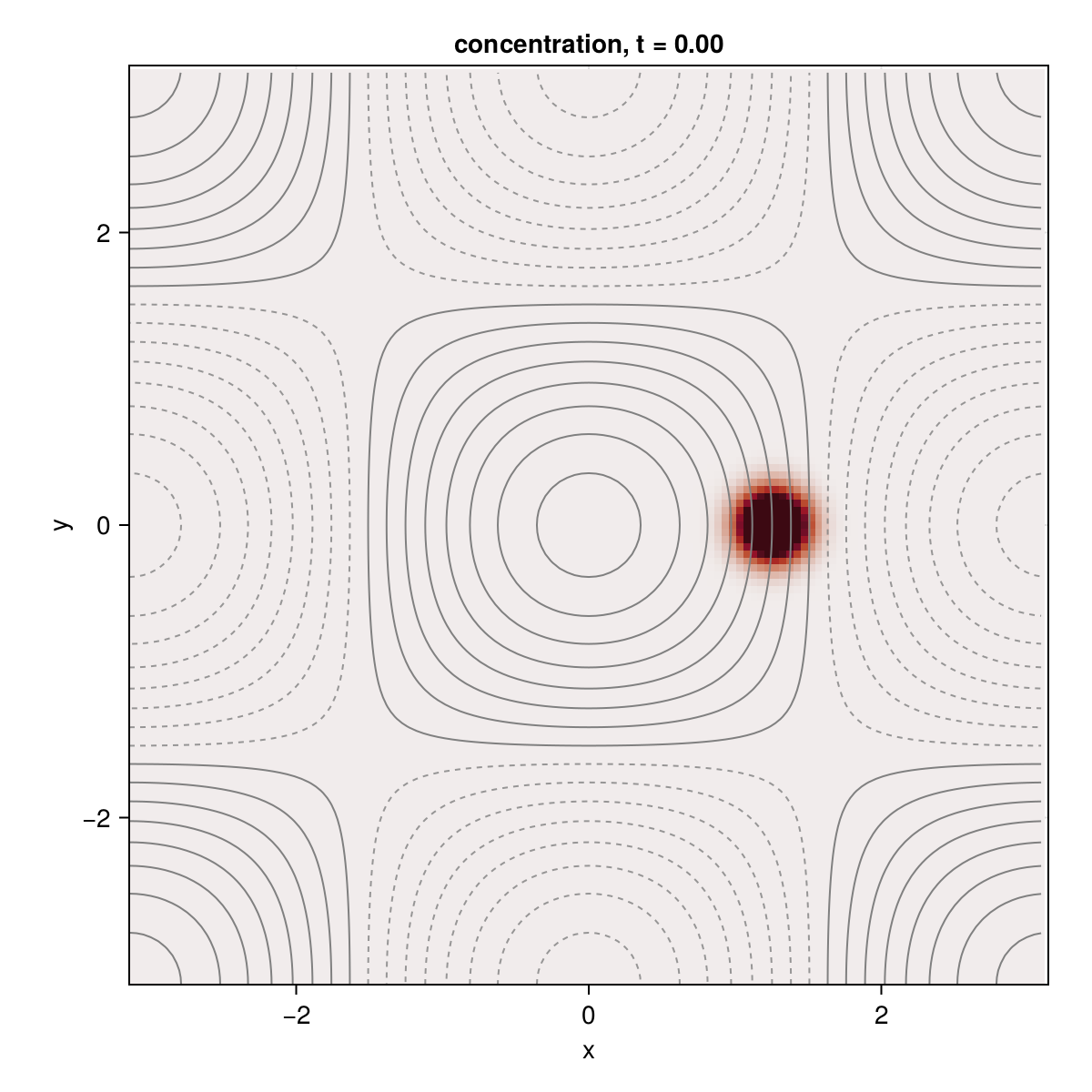
Now we time-step Problem and update the c_anim and title observables as we go to create an animation.
startwalltime = time()
frames = 0:round(Int, nsteps/nsubs)
record(fig, "cellularflow_advection-diffusion.mp4", frames, framerate = 12) do j
if j % (200 / nsubs) == 0
log = @sprintf("step: %04d, t: %d, walltime: %.2f min",
clock.step, clock.t, (time()-startwalltime)/60)
println(log)
end
c_anim[] = vars.c
title[] = @sprintf("concentration, t = %.2f", clock.t)
stepforward!(prob, nsubs)
TracerAdvectionDiffusion.updatevars!(prob)
end"cellularflow_advection-diffusion.mp4"This page was generated using Literate.jl.